nichemedical.net.au
ARTICLE IN PRESS The Journal of Emergency Medicine, Vol. xx, No. x, pp. xxx, 2009Copyright © 2009 Published by Elsevier Inc. doi:10.1016/j.jemermed.2008.06.029 Original Contributions EFFICACY AND COST COMPARISONS OF BRONCHODILATATOR ADMINISTRATION BETWEEN METERED DOSE INHALERS WITH DISPOSABLE SPACERS AND NEBULIZERS FOR ACUTE ASTHMA IN AN INNER-CITY ADULT POPULATION Su
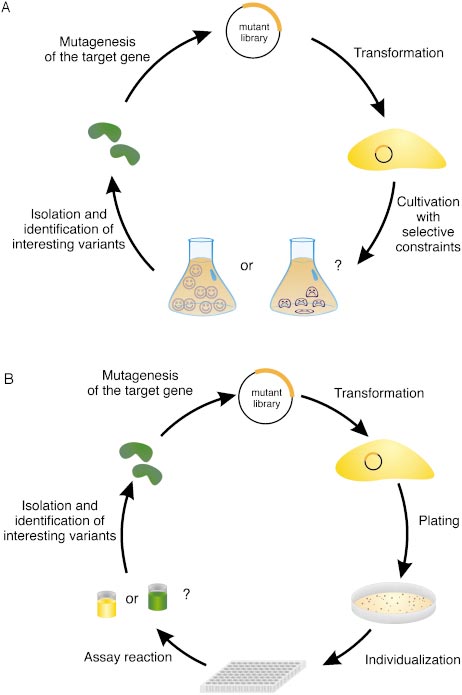 extremely rare. Codon-level random mutagenesis ofcomplete genes therefore would be desirable, but hasnot been realized yet.
extremely rare. Codon-level random mutagenesis ofcomplete genes therefore would be desirable, but hasnot been realized yet.